library(dplyr)
library(ggplot2)
library(tidyplate)
library(drc)
library(ggsci)Introduction
Here is an example of how to fit and analyse ELISA data using ggplot2, drc, and broom.
First we import plate data using the tidy_plate function from the tidyplate package:
raw_data <- tidy_plate("data/elisa_example.xlsx")
glimpse(raw_data)Rows: 50
Columns: 10
$ well <chr> "A01", "A02", "A03", "A04", "A05", "A06", "B01", "…
$ coat_protein_name <chr> "sBACE", "sBACE", "sBACE", "sBACE", "sBACE", "sBAC…
$ coat_protein_ug <dbl> 0.2, 0.2, 0.2, 0.2, 0.2, 0.2, 0.2, 0.2, 0.2, 0.2, …
$ coat_protein_source <chr> "NS0", "NS0", "NS0", "NS0", "NS0", "NS0", "NS0", "…
$ primary_mab_name <chr> "aBACE", "aBACE", "aBACE", "aSARS", "aSARS", "aSAR…
$ primary_mab_clone <chr> "6626.1", "6626.1", "6626.1", "CC6.29", "CC6.29", …
$ primary_mab_conc <dbl> 10.0000000, 10.0000000, 10.0000000, 10.0000000, 10…
$ secondary_mab_name <chr> "goat-aHuman", "goat-aHuman", "goat-aHuman", "goat…
$ secondary_mab_dil <chr> "1to5000", "1to5000", "1to5000", "1to5000", "1to50…
$ od450 <dbl> 1.396, 1.170, 1.299, 1.324, 1.170, 1.299, 1.374, 1…Prepare data before plotting
blank_data <- raw_data |>
filter(primary_mab_name == "blank")
mean_blank <- mean(blank_data[["od450"]], na.rm = TRUE)
summary <- raw_data |>
filter(primary_mab_name != "blank") |>
mutate(blanked_od = od450 - mean_blank) |>
group_by(primary_mab_name, primary_mab_conc) |>
summarise(
mean_od = mean(blanked_od, na.rm = TRUE),
mean_sd = sd(blanked_od, na.rm = TRUE),
.groups = 'drop'
)
head(summary)# A tibble: 6 × 4
primary_mab_name primary_mab_conc mean_od mean_sd
<chr> <dbl> <dbl> <dbl>
1 aBACE 0.00244 0.138 0.0797
2 aBACE 0.00977 0.214 0.0626
3 aBACE 0.0391 0.395 0.0644
4 aBACE 0.156 0.740 0.0531
5 aBACE 0.625 1.11 0.0358
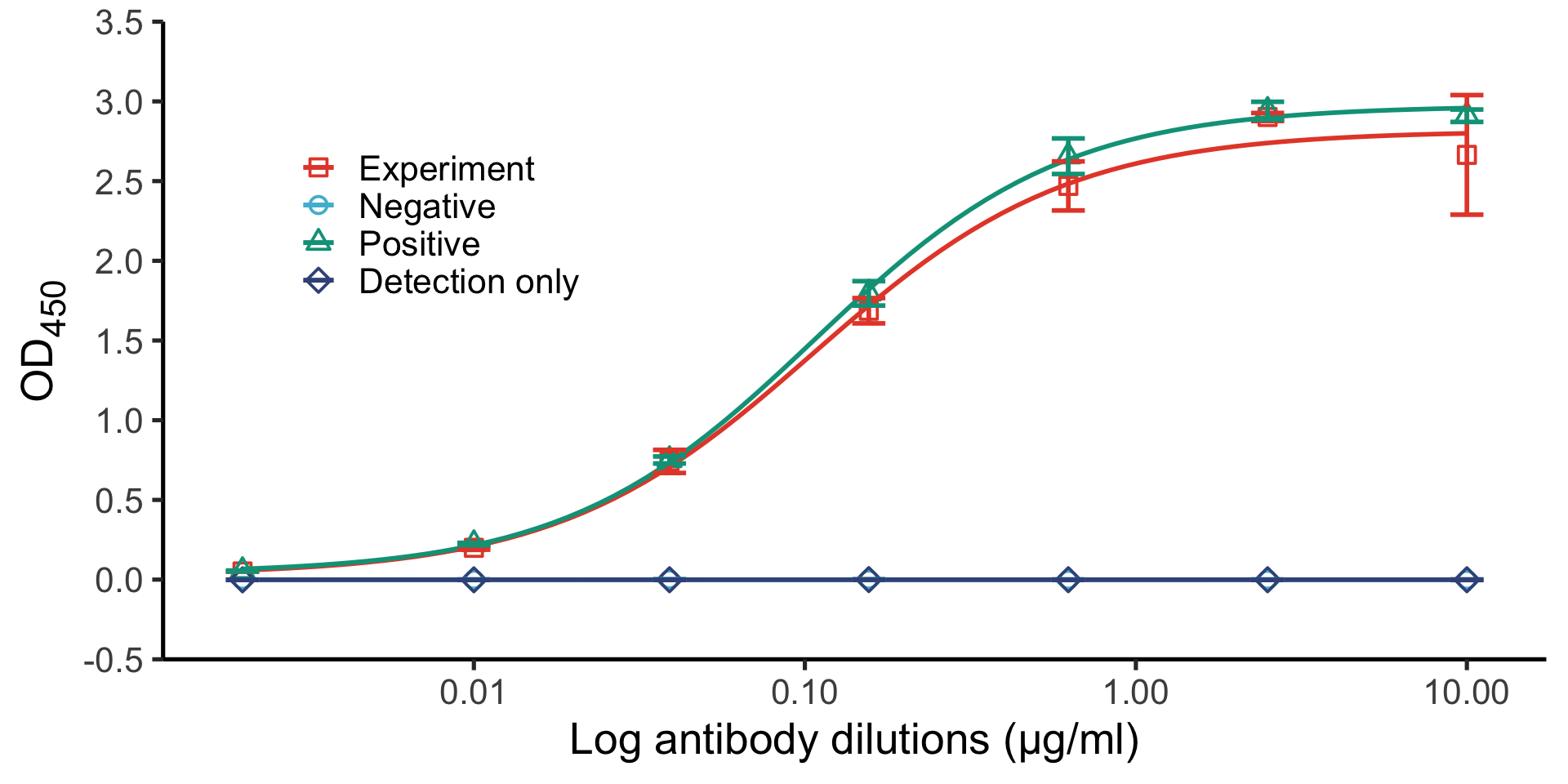
6 aBACE 2.5 1.28 0.0486The final plot
theme_set(theme_bw(base_size = 20))
ggplot(summary, aes(x = primary_mab_conc,
y = mean_od,
group = primary_mab_name,
color = primary_mab_name,
shape = primary_mab_name)) +
geom_point(size = 3, stroke = 1.5, show.legend = TRUE) +
geom_smooth(method = drc::drm,
method.args = list(fct = drc::L.4()),
se = FALSE, linewidth = 1,
show.legend = FALSE) +
geom_errorbar(aes(ymin = mean_od - mean_sd,
ymax = mean_od + mean_sd),
width = 0.1, show.legend = FALSE) +
scale_x_log10() +
scale_color_npg(name = NULL) +
scale_shape(name = NULL) +
labs(x = "[Antibody] (Log)",
y = expression(OD[450])) +
theme(legend.position = c(0.15, 0.8),
legend.box.background = element_rect())